Privacy-preserving Sequence Alignment Map
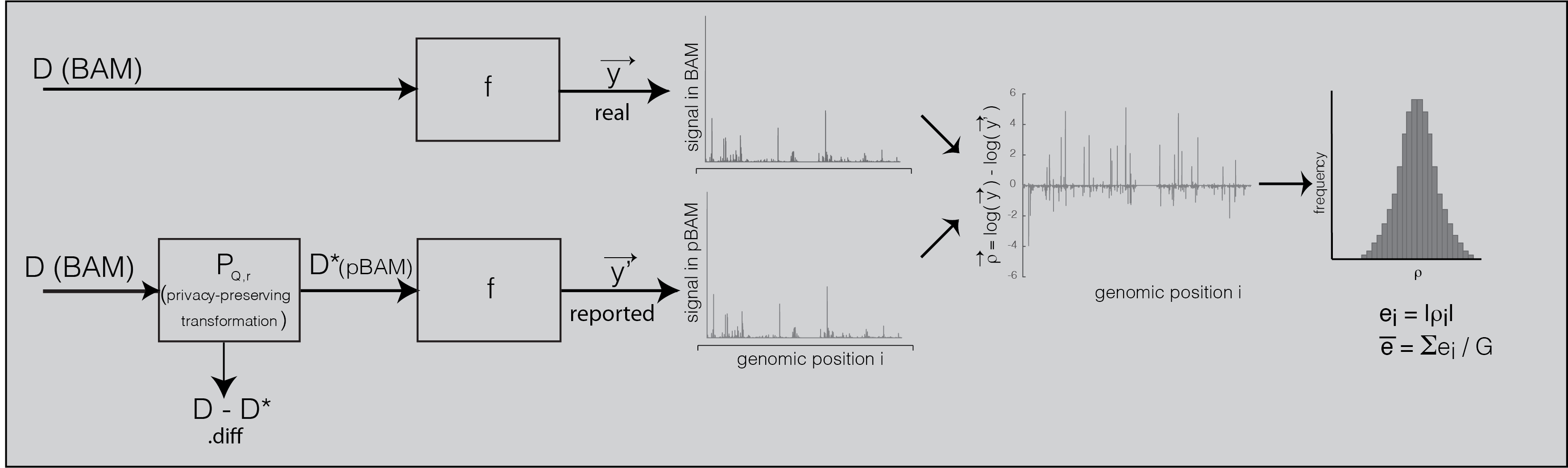
Privacy-preserving file format manipulation system has two components: (1) A publicly sharable alignment file (pBAM), (2) a private and light-weight file that needs secure access (.diff). We provide a combination of scripts called “pTools” that can convert alignment files into privacy-aware file format system and vice versa. pTools are implemented in bash, awk and Python. .diff files are encoded in a compressed format to save disk space. For convenience, pBAM files are saved as BAM files with manipulated content and with a p.bam extension. That is, any pipeline that uses BAM as an input can take p.bam as an input as well. For 10x single-cell RNA-Seq data, we have a module that converts pBAM files into pFastq format such that it can be used with pipelines that use fastq files as input. This file format manipulation has been adopted by the ENCODE Consortium Data Coordination Center and deployed in ENCODE Uniform Pipeline Framework using workflow description language (WDL) scripts, docker images, accompanied with appropriate documentation for computational reproducibility on multiple platforms (google cloud, slurm scheduler, LINUX servers, etc.) under ENCODE Data Processing pipelines. pTools can be accessed from download page and ENCODE DCC GitHub page. Examples and file specifications of BAM, pBAM, pCRAM can be found at the documentation page.
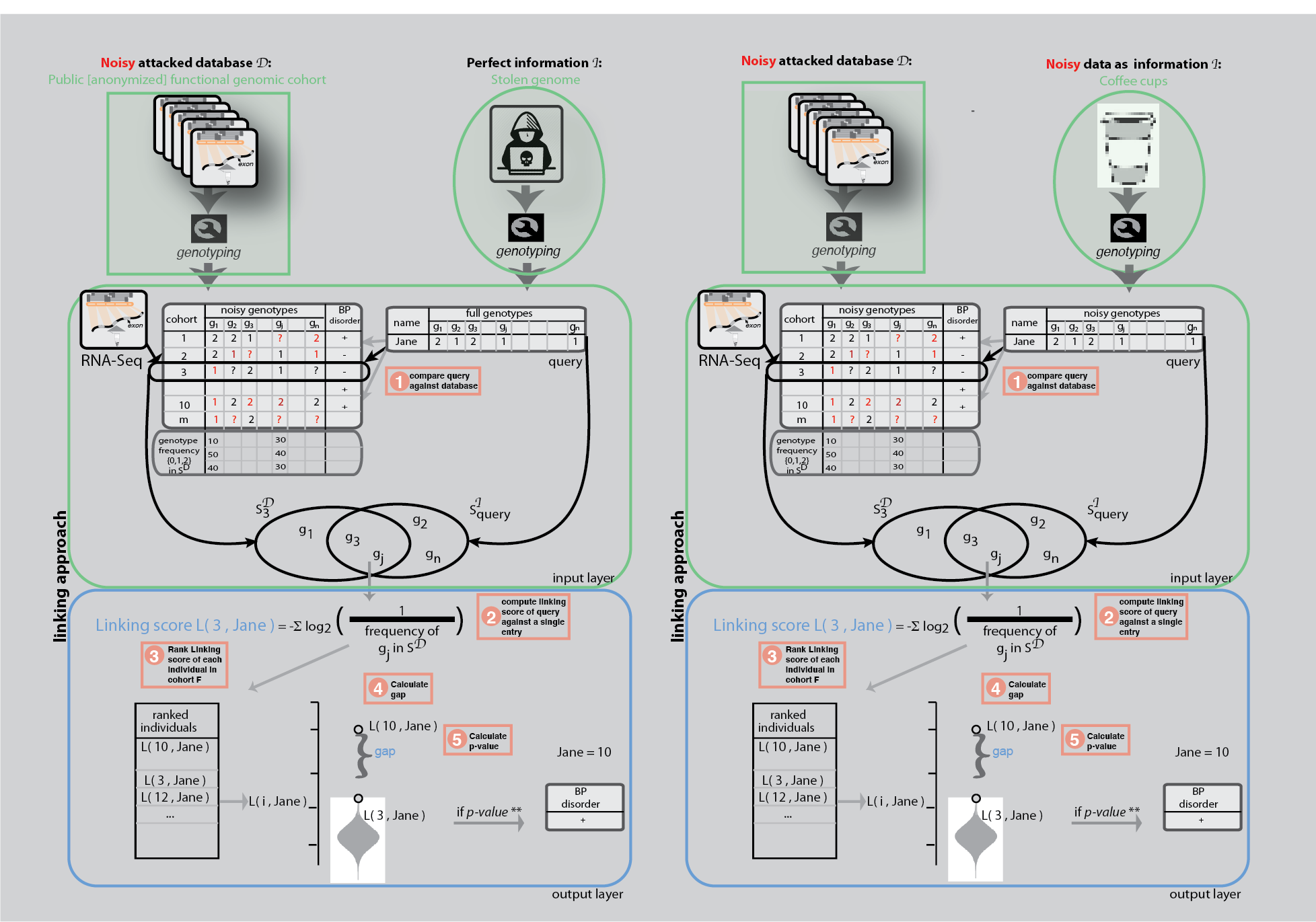
Linkage attacks
Linkage attacks have been shown to be powerful statistical tools to understand the private information leakage in a dataset. A famous example of a linkage attack is the de-anonymization of Netflix Prize challenge data using IMDB user comments [1]. Within genomics, the genetic information in genealogy websites have been overlapped with short tandem repeats on the Y chromosomes to infer the surnames of participants in genomics datasets [2]; and eQTL data have been shown to be able to de-anonymize gene expression datasets [3,4]. Linkage techniques have been used outside of a privacy context as well, for example, to resolve sample swap problems during omics data production.
We used linkage attacks first to show how sensitive phenotypes can be inferred from anonymized functional genomics dataset when we have the full genome of a known individual. This was somewhat obvious as it was previously shown that a great number of genotypes can be obtained from functional genomics reads.
We then showed a not-so-obvious linkage attack by using used coffee cups to mimic a real world hacking scenario. We extracted DNA from the surface of coffee cup lids and performed low coverage sequencing. We showed that a linkage between an anonymized RNA-Seq data and a coffee cup sample is possible as low as $19 cost.
[1] Narayanan A, Shmatikov V. Robust De-anonymization of Large Sparse Datasets in Proceedings of 2008 IEEE Symposium on Security and Privacy (sp 2008), 2008:111-125.
[2] Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science, 2013;339(6117):321-324.
[3] Schadt EE, Woo S, Hao K. Bayesian method to predict individual SNP genotypes from gene expression data. Science, 2012;44(5):603-608.
[4] Harmanci A, Gerstein M. Quantification of private information leakage from phenotype-genotype data: linking attacks. Nature Methods, 2016;13(3):251-256.